CRDS Scan Analyzer
Basics
-
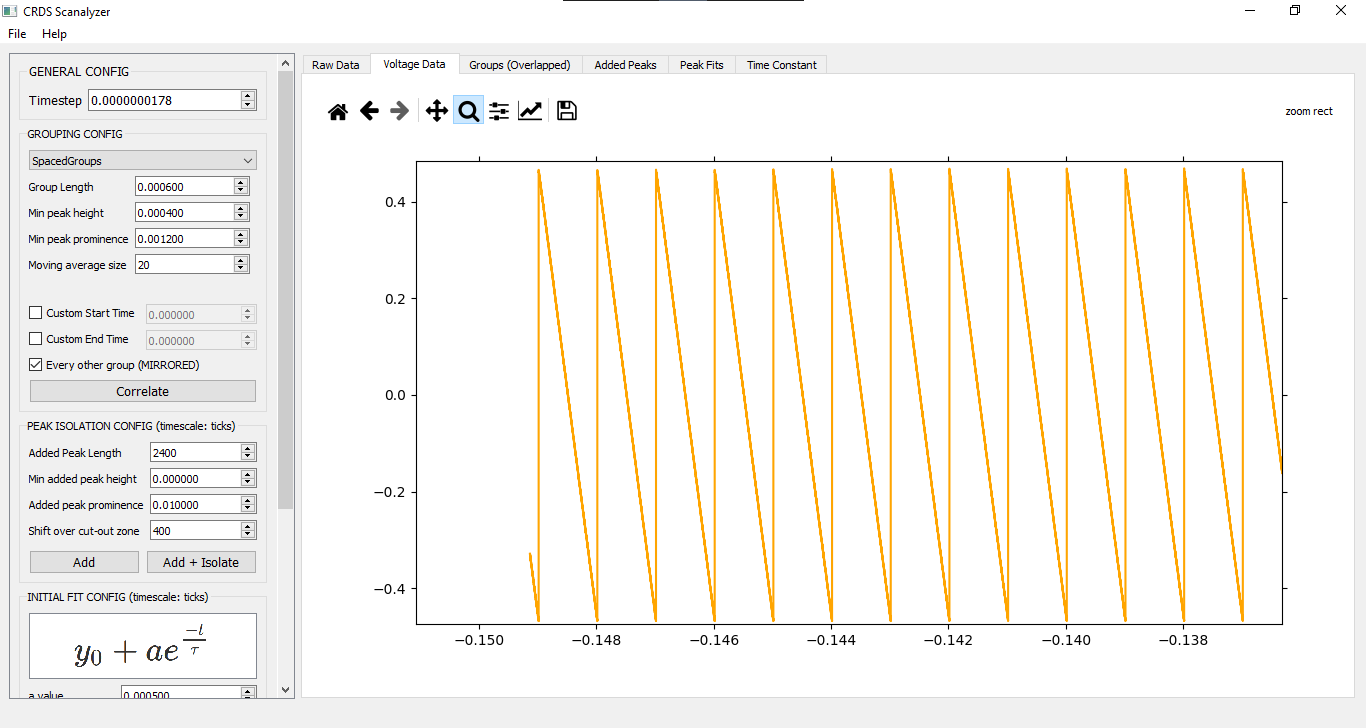
Designed for usage with Mid-IR laser comb scan output (time scale, not frequency)
-
Expected data column format:
<time>, <signal voltage in>, <piezo crystal voltage out (optional)>
Building (Python 3.8+)
-
pip3 <or python3 -m pip> install -r requirements.txt -
build.cmd -compileUI -build -
Find build output in
/dist/
General Usage
-
Import data (CSV - default Zurich output format)
- Comment - %
- Delimiter - ;
-
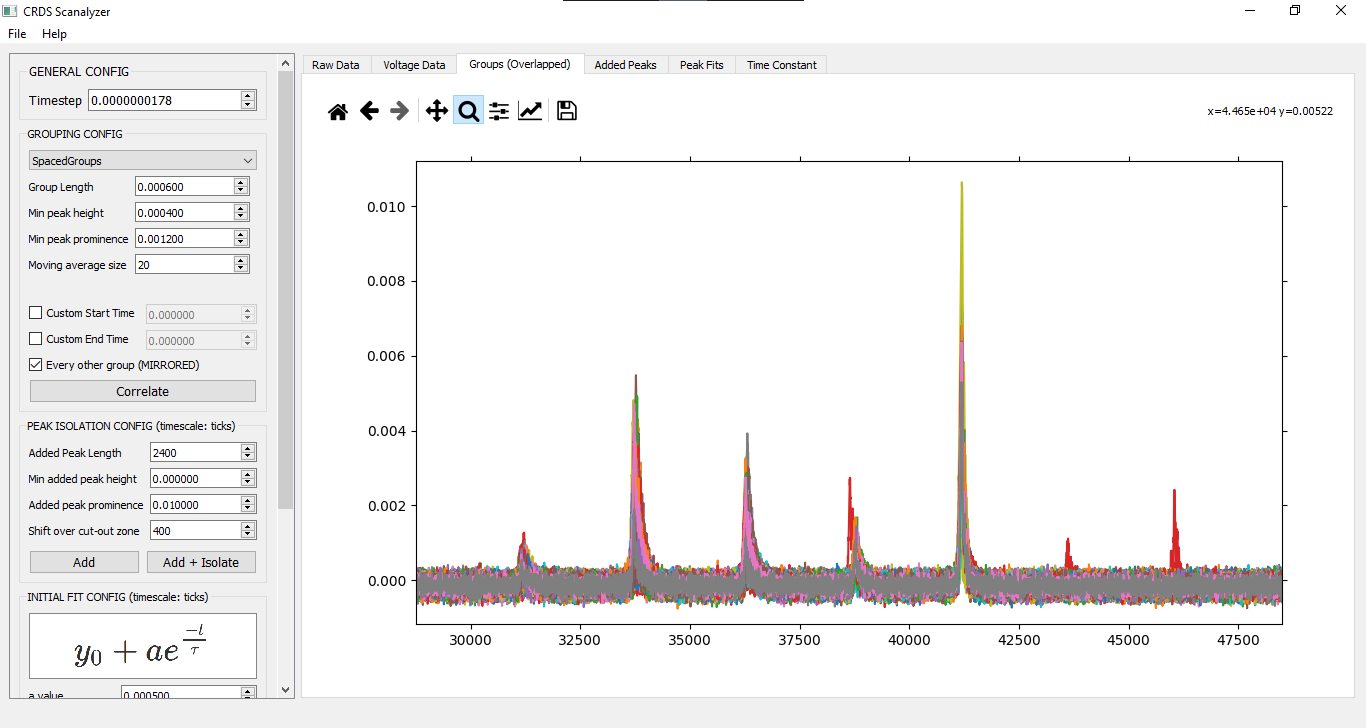
Cut out & overlay peak groups using either piezo-voltage-threshold or group-spacing algorithm
-
Set peak isolation parameters + guesses for ringdown function coefficients
-
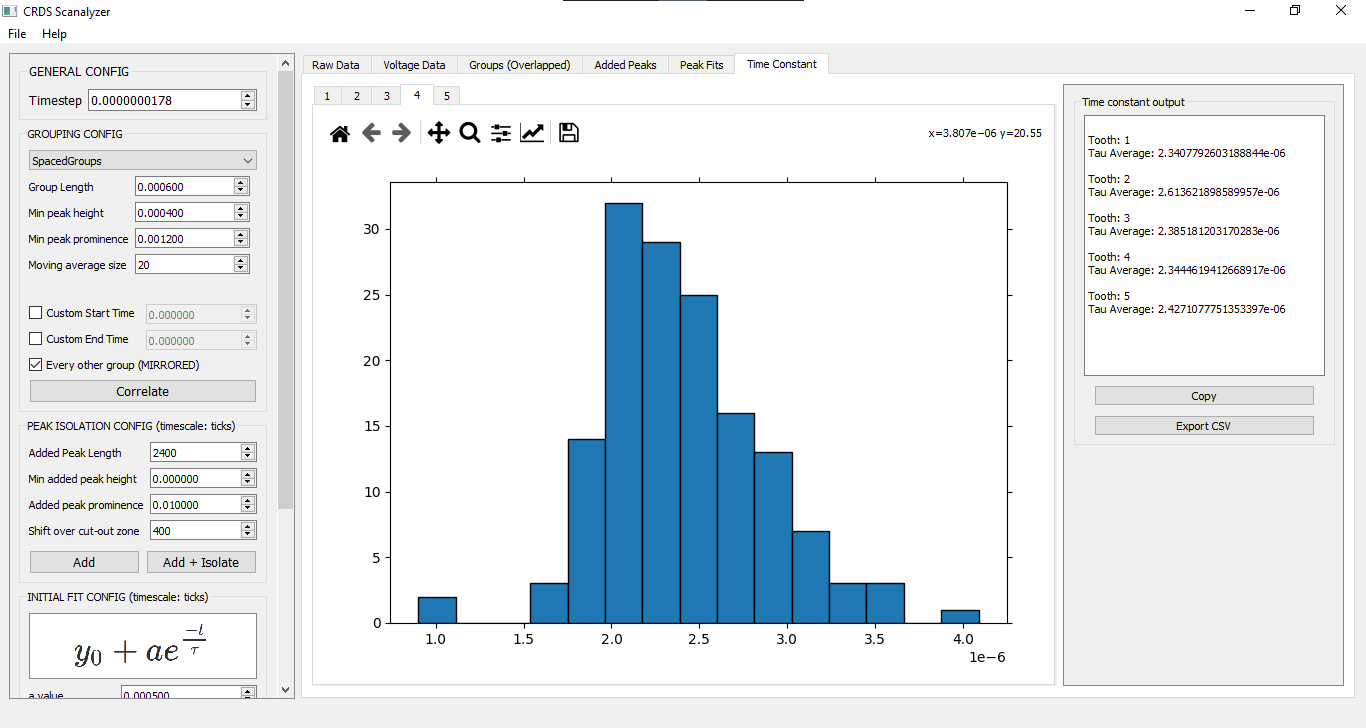
Admire glorious tau distributions for each comb tooth - Note that the timescale used during correlation, adding & fitting (including the initial guess for tau) is in SENSOR TICKS. The resulting tau values are converted back to the original timescale at the end.
Instructions
GENERAL CONFIG
| Input | Purpose |
|---|---|
| Timestep | Time interval of each sensor tick (e.g. 0.0000000178s) |
GROUPING CONFIG (SpacedGroups)
The SpacedGroups algorithm uses the length of each group (looking length in each direction from the first peak in the group) to determine which peaks fit into which group.
| Input | Purpose |
|---|---|
| Group length | How long in time the peak group is (e.g. 0.0006s) |
| Min peak height | The lowest acceptable height for a peak to be detected |
| Min peak prominence | Prominence of detectable peaks - read about prominence here. Is finnicky, might have to play around a bit |
| Moving average size | Peak detection uses a moving average (of which this is the denominator) to smooth out the data (FOR GROUPING ONLY). |
Notes
-
You can set custom start and end cutoffs for grouping in case there's only part of a group at the beginning or end
-
You can choose to exclude mirrored groups (which occur every other group). Or don't. Depends on how you configured the lab setup.
GROUPING CONFIG (VThreshold)
This algorithm should look at the min and max voltages provided to separate out groups. It's not implemented yet, though.
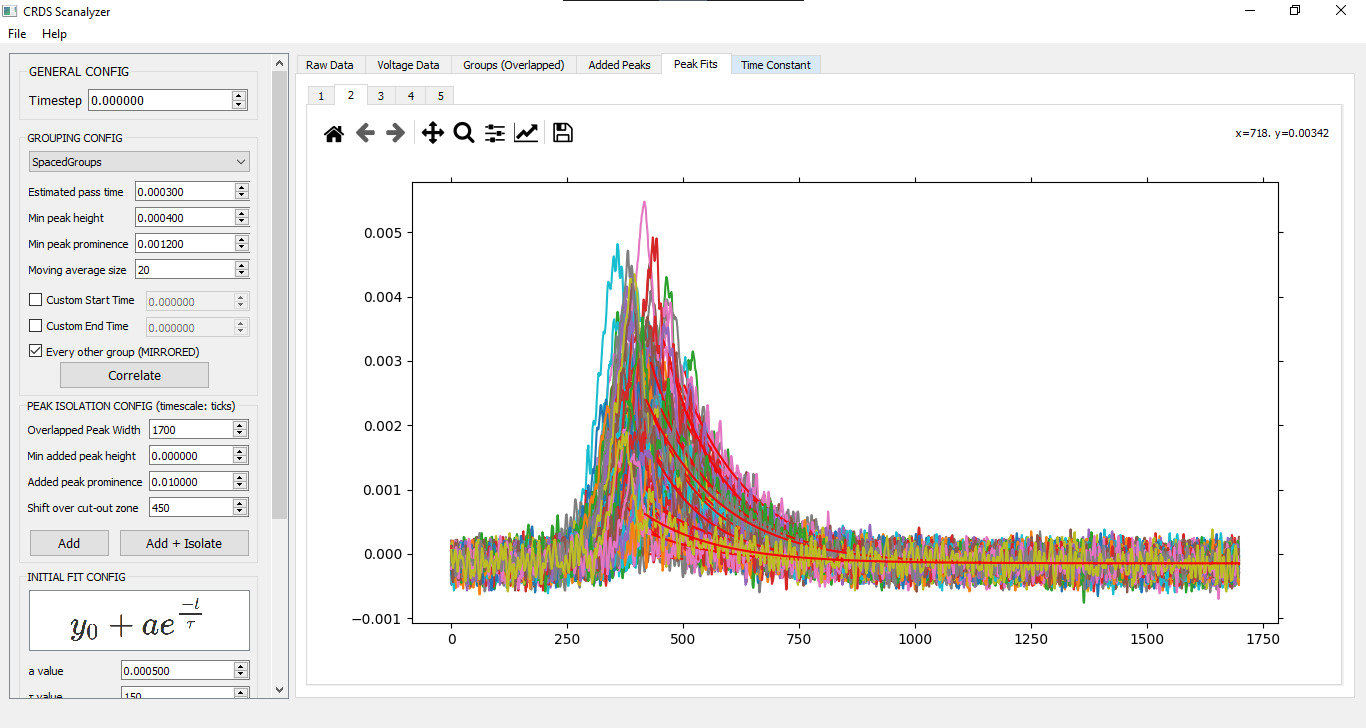
PEAK ISOLATION CONFIG
This is where the overlayed peak groups are added together, and the added peaks are cut out.
Note that the timescale here is in ticks, NOT seconds/milliseconds/etc.
| Input | Purpose |
|---|---|
| Added peak length | The length of the (added) peaks that you want cut out. Centers around tip of peak. |
| Min added peak height | The lowest acceptable height for a peak to be detected |
| Added peak prominence | Prominence of detectable peaks - read about prominence here |
| Shift over cut-out zone | How many ticks to shift the center of the cut-out zone from the peak tip |
INITIAL FIT CONFIG
y = y0 + a*e^(-t/tau)
| Input | Purpose |
|---|---|
| a value | Variable in equation |
| tau value | Variable in equation |
| y0 value | Variable in equation |
| Shift over fit start | How many ticks to shift over the start of the curve fit |
Feature Set
| Feature | Status |
|---|---|
| Import CSV Data | ✅ |
| Group peaks (peak spacing) | ✅ |
| Group peaks (piezo crystal voltage thresholds) | ❌ |
| Custom start & end times for peak grouping | ✅ |
| Correlate grouped peaks | ✅ |
| Add & cut out overlayed peaks | ✅ |
| Batch fit peaks | ✅ |
| Display average tau data | ✅ |
| Export data as CSV | ✅ |
| Residuals on fits | ✅ |
| Standard deviations | ✅ |
| Persistent data storage | ❌ |
Ideas if anyone has the time to add anything
-
Speedier iteration & number crunching (Numba)
-
Support for different file formats (e.g. [LabView](npTDMS · PyPI))
Screenshots